Nanopore Protein Sequencing
by: Abbas Zaki
Biotech Breakthrough: Nanopore Protein Sequencing
Nearly 20 years after the completion of The Human Genome project, DNA sequencing technology has become a core component of biomedical research. The development of DNA sequencing techniques such as bridge amplification paired with sequencing by synthesis has afforded incredible insight into not just the genome but the epigenome and transcriptome as well. However, 10 years after the beginning of the Human Proteome Project, probing the proteome has remained elusive.
Current Technology Limitations: Mass Spectrometry
Being able to quantify every protein in a cell and to determine what post-translational modifications are present would be invaluable in understanding disease mechanisms, progression, signaling pathways and much more. While a powerful technique, protein sequencing by mass spectrometry typically requires picomolar amounts of samples thereby limiting the detection of low-abundance proteins which might be present in as few as 10-100 molecules/cell [1]. Enrichment of rarer proteins using antibodies is possible in some circumstances; however, prior knowledge of the proteins of interest is usually needed [1]. As such, discovery of rare and low-abundance proteins is not typically feasible with mass spectrometry. Mass spectrometry has also not been able to provide throughput comparable to DNA sequencing technologies and has limited capabilities when dealing with longer amino acid sequences [2]. Nanopore sequencing, a more recent approach for DNA sequencing offers a potential way to sequence proteins de-novo at a single molecule resolution.
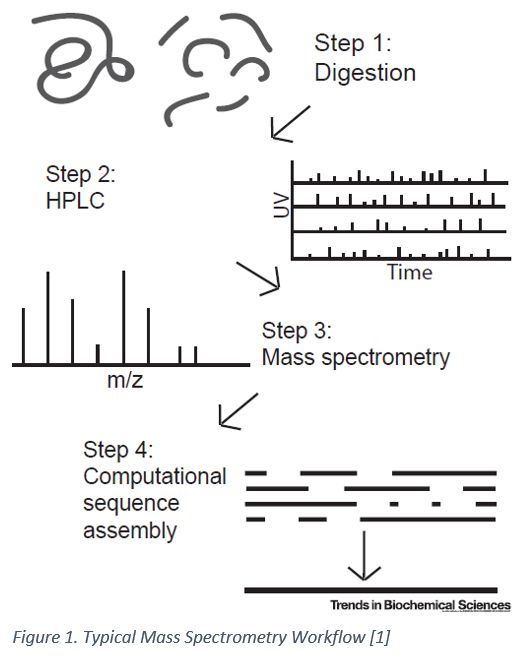
Nanopore Sequencing: An Overview
The key components of a Nanopore Sequencing device are two fluidic compartments separated by a membrane (typically a lipid bilayer) with proteins known as nanopores inserted in the membrane. Electrodes connected to each compartment create a voltage difference that results in a current measured across the nanopores as ions in the solution transit through the nanopore [2]. While ionic components of the solution as chloride, potassium, sodium, etc. are relatively small compared to the pore and can transit freely, a larger molecule occludes more of the volume within the nanopore, creating in resistance to charge movement across the nanopore that can be detected as a change in the current being measured. Coupling nanopores with helicases and polymerases for DNA sequencing results in a controlled translocation of each nucleotide across the nanopore such that stepwise changes in the current can be detected at predictable rates to identify the nucleotide currently occupying the nanopore. Nanopores have been successfully commercialized by Oxford Nanopore Technologies (ONT) for genomic sequencing and multiple groups have developed methods to uncover epigenetic and transcriptomic information using nanopores as well. Since nanopore sequencing measures the current change resulting from a molecule occupying the nanopore, many groups have hypothesized that a properly sized nanopore might be able to sequence amino acids in much the same way as nucleotides.

Challenges with Nanopore-Based Protein Sequencing:
However, nanopore-based protein sequencing faces some key challenges. Unlike 4 nucleotides that make up DNA, proteins are composed of 20 amino acids, each with a different charge and virtually 100s of potential post-translational modifications [2]. Moreover, it has proved difficult to control the translocation of peptides across a nanopore as enzymes capable of ratcheting the peptide at a controlled rate have been difficult to identify and conjugate with existing nanopores. The first issue can be solved at least in part by computational approaches. Due to the high repetition of some amino acids in most peptides, it has been estimated that 97.5% of the proteome can be identified by reading just 3 key amino acids robustly in a 50 amino acid fragment [3]. Therefore, the main challenges to nanopore-based protein sequencing are engineering pores with the correct geometry for amino acids and controlling the translocation of peptides across these pores.
A Plan Unfolds: Proteasomes as A Potential Solution?
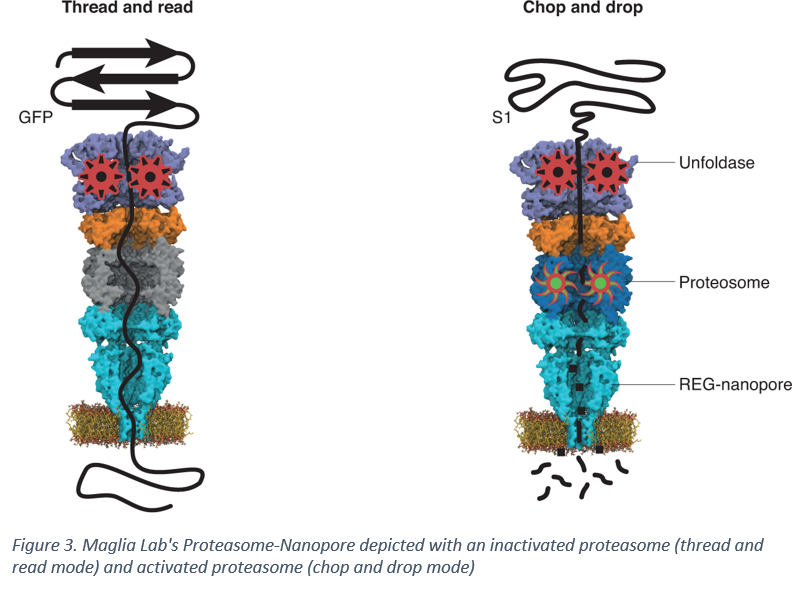
Giovanni Maglia, a Professor of Chemical Biology at the University of Groningen in the Netherlands recently engineered a multi-component proteosome-nanopore that holds potential to be the missing link needed to translocate peptides in a controlled manner across a nanopore. Maglia Lab's work was featured in a Nature Chemistry article in November last year and provides a method for bottom-up fabrication of a multi-protein complex capable of unfolding and threading proteins in a controlled manner. Dr. Maglia's designed a synthetic nanopore that paired proteosome activator 28α (also referred to as REG or 11S activator) from mice to the anthrax protective antigen nanopore from Bacillus Anthracis by replacing the disorder region of REG with the β-barrel transmembrane region of the nanopore [4]. Since REG regulates the function of the proteosome, it was fused to 20S proteosome from T. acidophilum to generate a proteosome-nanopore [4]. This proteosome-nanopore allows for unfolding the peptide via a vasolin-containing protein-like ATPase in the 20S proteasome and either threading the peptide through the nanopore (if the proteasome is inactivated by mutating a residue in the proteolytic site) or chopping the peptide into smaller fragments that can be dropped into the nanopore and sequenced [4].
The work by Maglia lab is a major step forward in enabling nanopore-based protein sequencing. While the current version of the nanopore can not distinguish the electrical signature of the amino acids with sufficient certainty to accurately determine the sequence, the ability to thread a peptide through a nanopore is a major achievement in the field and opens the door for coupling with modified nanopores that are more sensitive to the translocation of individual amino acids such that each amino acid can be accurately labelled.
Future Outlook:
A proteasome-nanopore like the one developed by Dr. Maglia's group provides a critical piece of the puzzle, leaving the optimization of nanopore geometries for amino acids as the main ongoing challenge in sequencing proteins de novo. Many groups are already working on the problem and the field of DNA sequencing has paved much of the way for engineering proteins to obtain optimized nanopores. Therefore, it is possible that an optimized nanopore integrating the thread-and-read mechanism described in Dr. Maglia's work could soon be developed. If the $4 billion market for DNA sequencing [5] is a predictor for the growth possible in protein sequencing, there exists significant incentive for developing an integrated nanopore-based solution capable of detecting and quantifying the vast underexplored landscape of the proteome.
Sources:
[1] Callahan, N., Tullman, J., Kelman, Z. & Marino, J. Strategies for Development of a Next-Generation Protein Sequencing Platform. Trends in Biochemical Sciences 45, 76-89 (2020).
[2] Oldach, L. The scramble for protein nanopore sequencing. Asbmb.org (2022). at <https://www.asbmb.org/asbmb-today/science/042722/the-scramble-for-protein-nanopore-sequencing>
[3] Palmblad, M. Theoretical Considerations for Next-Generation Proteomics. Journal of Proteome Research 20, 3395-3399 (2021).
[4] Zhang, S., Huang, G., Versloot, R.C.A. et al. Bottom-up fabrication of a proteasome-nanopore that unravels and processes single proteins. Nat. Chem. 13, 1192-1199 (2021). https://doi.org/10.1038/s41557-021-00824-w
[5] DNA Sequencing Market Share & Growth Report, 2020-2027. Grandviewresearch.com at <https://www.grandviewresearch.com/industry-analysis/dna-sequencing-market#:~:text=The%20global%20DNA%20sequencing%20market,the%20identification%20of%20DNA%20variations.>

